Marine Threat: Invasive Species
Marine Threat: Invasive Species
Consultant: Maciej Maselko
Academic & Industry Insight
John Teem (International Life Sciences Institute)
Owain Edwards (CSIRO)
Pictured Above: The Crown of Thorns Starfish preys upon corals, and its predation plays an important role in maintaining species diversity. However, high-density outbreaks are occurring with more frequency as a result of climate change, leading to widespread damage of reefs.
BACKGROUND
Invasive species are non-native organisms that have been introduced into a new ecosystem (Richardson et al. 2000). A species irruption is a sudden change in the population density of an organism, typically characterized by population explosions followed by subsequent crashes (Roughgarden 1996). Both invasives and native-species irruptions can cause harmful impacts to natural resources and/or human use of the resource (Bax et al., 2003; Molnar et al., 2008). The eradication and control of such species can be particularly challenging in marine environments, and managers often rely on time- and labor-intensive manual removal (Sutherland et al., 2017).
Invasions and irruptions can displace native species (e.g. Huxel 1999), cause the loss of native genotypes (e.g. Gurevitch & Padilla 2004), lead to a reduction in biodiversity, modify habitats, change community structure, affect food-web properties and ecosystem processes (Byrnes et al. 2007; D’eath et al. 2014), impede the provision of ecosystem services (e.g. Katsanevakis et al., 2014), impact human health (Pyšek & Richardson 2010), and cause substantial economic harm (Marbuagh et al. 2014). Pimentel and Colleagues (2005) estimate invasive species in the United States cause major environmental damage, as well as economic losses totaling $120 billion per year.
Rapid globalization and increasing trends of trade, travel, and transport in recent decades have accelerated marine biological invasions by increasing rates of new introductions through various pathways, such as shipping, navigational canals, aquaculture, and the aquarium trade (Molnar et al. 2008; Riccardi et al. 2017). At any given moment an estimated 10,000 different species are being transported in ballast tanks alone (Carlton 2001). Furthermore, climate change is predicted to cause significant changes in species ranges and habitats, altering pathways for migration, and increasing flooding, thus also increasing the frequency and extent of species invasions (Rahel & Olden 2008).
Harmful invasive/irruptive species that damage ocean ecosystems include:
- The crown of thorns starfish (Acanthaster planci) is native in the Indo-Pacific region and is not harmful at low population densities. However, irruptive crown of thorns starfish (COTS) population booms are considered as a significant threat to coral reefs and are responsible for approximately 42 percent of observed cover loss in the Great Barrier Reef during an irruption (D’eath et al. 2014). Localized nutrient pollution can lead to an increased abundance of COTS and increased sea temperatures is expected to also increase both abundance and range of COTS (Uthicke et al. 2015).
- The Northern Pacific Sea Star (Asterias amurensis) is native to the North Pacific along the coasts of Russia, China, and the Korean peninsula. This sea star is a problematic invasive along the south of Australia. During large population booms, the sea star voraciously consumes endangered native species (Ross et al. 2004).
- Purple sea urchins (Strongylocentortus purpuratus) are native along the Pacific Coast of the United States. Populations of the purple sea urchin’s natural predators, such as sea otters and sea stars, have dwindled (Pearse 2006). The resulting purple sea urchin population explosion has decimated kelp forests which are critical habitats for many marine species (Pearse 2006).
- Lionfish (Pterois volitans and P. Miles) are native to the South Pacific and Indian Oceans. Along the Eastern Coast of the United States and in the Caribbean, non-native lionfish are voracious fish predators and cause damage to coral ecosystems and native fish populations (Ballew et al. 2016).
- The European Green Crab (Carcinus maenas) is native to Europe and North Africa. Invasive in the Americas, Australia, and South Africa, the crab preys on a wide variety of organisms, but is particularly harmful to bivalves (Colnar & Landis 2007).
- The American Blue Crab (Callinectes sapidus) is native to the American Atlantic. As an invasive species carried east by transatlantic trade, it has regularly been observed on the Atlantic coast of Europe from Denmark to Portugal, as well as along the western and southwestern shores of the Mediterranean. An omnivore with high trophic flexibility, the American Blue Crab is also aggressive, a fast grower, and has a short reproductive cycle. As a result, invasive populations have expanded exponentially, wiping out native species (Fuentes et al. 2019).
PROGRESS TO DATE
Detection and Prevention
eDNA provides information on species presence without the need for capture or direct observation. eDNA is being used to study invasions in freshwater environments. eDNA was used for the first time to confirm the presence of a freshwater aquatic invasive species, the American bullfrog (Rana catesbiana), in 2008 (Ficetola et al. 2008), and has subsequently been used to detect other freshwater invasive species in a variety of environments, such as Mozambique tilapia (Oreochromis mossambicus) in the tropics (Robson et al. 2013) and Asian carp in the American Great Lakes (Jerde et al. 2011). Scientists and managers predict that in short order, rapid growth, widespread deployment, and automation of eDNA techniques will transform the sensitivity, speed, and scale with which we detect alien species (Riccardi et al. 2017). Detection of invasive species in the ballast water of incoming vessels at ports could be a strategic focal point of monitoring efforts (Zaiko et al. 2015).
Biocontrol
Current underwater control efforts of marine invasive and irruptive species primarily involve killing or capture by divers. These methods have been relatively successful for small geographic areas. For example, COTS are removed by divers or injected with chemicals (e.g. acetic acid). Targeted trapping has shown some promise for green crab population control (Duncombe & Theriault 2017). Generating a market to drive harvest of invasive such as the edible European Green Crab are underway. Lionfish is also edible and has an emerging commercial harvest, despite the fact that they can be toxic to humans (Morris et al. 2012). While these efforts have had some success, they are expensive and labor-intensive, and are difficult to scale.
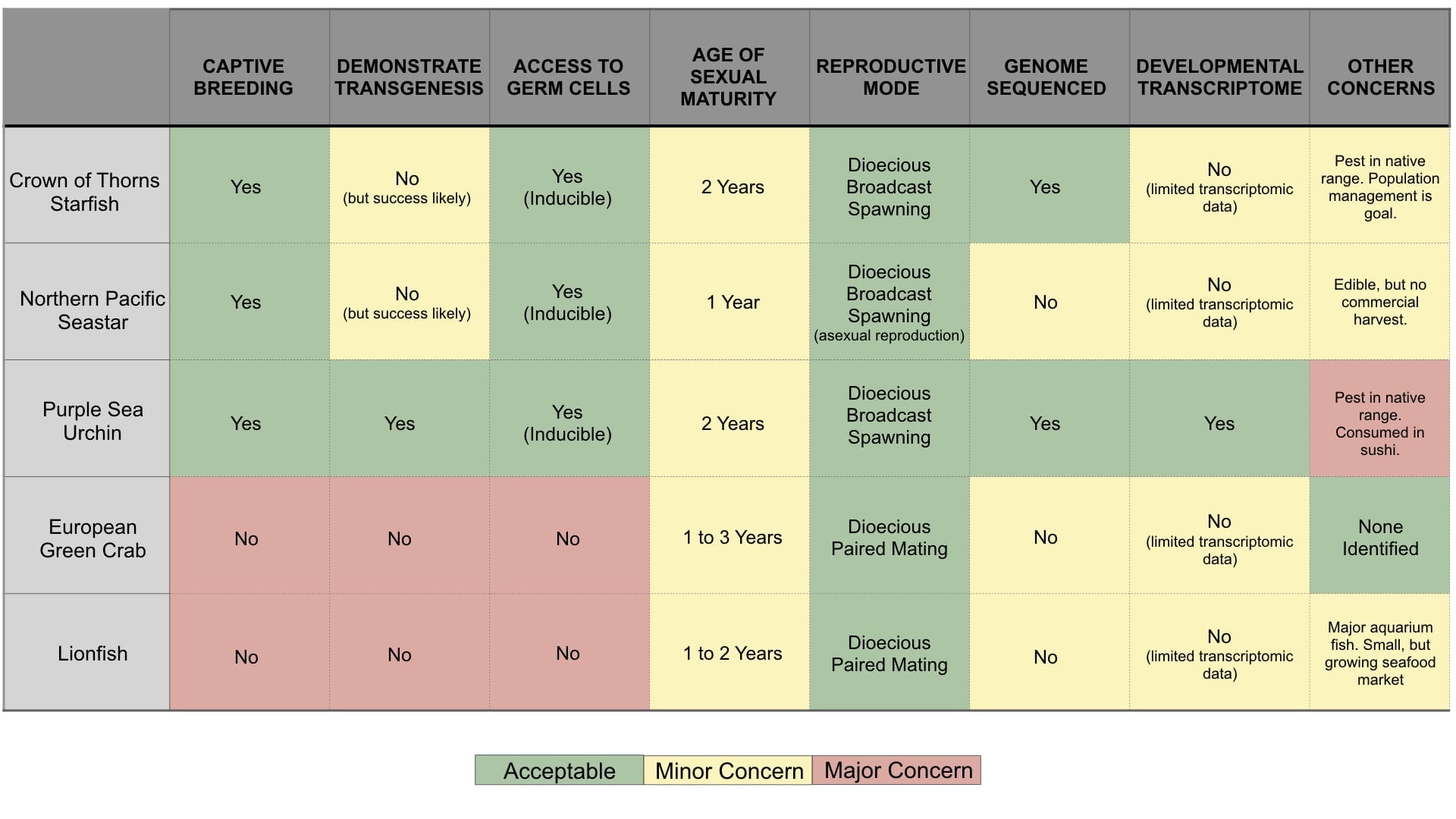
There is a varying degree of progress to date with respect to understanding the genomics of the most significant invasive and irruptive species in the ocean. Table 1 presents a summary of the current state of the art including progress of breeding and genomics knowledge as well as special considerations such as the potential for social acceptance of genetic biocontrol. Based on this summary information, COTS and Northern Pacific Sea Stars (Asterias amurens) seem the most likely potential candidates for genetic biocontrol.
- COTS will spawn and breed in captivity and reach sexual maturity in two years, (Kessing et al. 1997). These factors are conducive to genetic manipulation. The genome has been sequenced, and potential aggregating and alarm pheromones identified, which open the door to genomic approaches for disrupting mating behavior (Hall et al. 2017). The primary technical hurdles to genetic manipulation efforts are establishing transgenesis protocols, and developing molecular components such as promoters and terminators.
- Northern Pacific Sea Stars breed in lab environments where it is possible to artificially induce spawning (Kanthani 1969). As with COTS, the primary technical hurdles to genetic manipulation are establishing transgenesis protocols and developing molecular components.
- Purple Sea Urchins are a model species for embryology with a long history of lab breeding, making genetic manipulation very feasible. The genome has been sequenced (Sodergren et al., 2006), spawning can be induced (Stepicheva & Song 2014), and transgenesis (Rast, 2000) and genome editing (Lin & Su 2016) demonstrated. The primary technical hurdles are developing molecular components such as promoters and terminators. Another potential major drawback to genetic biocontrol of sea urchins is the market for their use in sushi which may lead to public resistance to transgenic animals in the wild.
- For Lionfish, genetic manipulation is not currently feasible because they have not yet been bred under laboratory conditions, despite efforts of Florida Tropical Aquaculture Lab (John Teem, personal communication). Further, consumer fear of eating transgenic lionfish may provoke resistance to genetic biocontrol and undermine complementary control efforts via commercial harvest.
- European Green Crab genetic manipulation is not currently feasible. Despite the fact that embryos can be collected from wild-caught crabs, they have not yet been bred in captivity.
INNOVATION
Chemical and physical methods as described above may be effective in reducing large problem species populations in a small area. However, they rarely eliminate the last few individuals which can subsequently repopulate the region. Genetic biocontrol methods have greater potential to control this residual population and contain unwanted and harmful species.
Genetic biocontrol is the release of organisms that have been genetically engineered for the purposes of controlling a pest species (Thresher et al. 2014). The engineered individual encounters and mates with the wild members of the species and introduces a genetic system that reduces the population size through a variety of mechanisms. An important benefit of genetic biocontrol is the potential to reduce pest species populations while minimizing the off-target effects. Other potential benefits include reduced toxicant use, more humane (non-lethal) approaches, and expanded application in locations where there may be human conflicts with other less specific methods (Campbell et al. 2015). Genetic biocontrol represents a potentially transformative advance for harmful ocean invasives that is not readily achievable with current technology (IUCN Synbio Assessment).
Repressible Lethal Systems
The Crown of Thorns Starfish and Northern Pacific Sea Star are feasible targets for genetic biocontrol through the use of repressible lethal disruption systems. Repressible lethal systems involve genetic circuits where immature life stages require the presence of a “repressor” molecule (often tetracycline) that prevents a toxic gene from getting expressed. Mature organisms no longer need the repressor and can be released. Offspring resulting from crosses between wild and engineered organisms die in the absence of the repressor (Thomas et al., 2000). The Commonwealth Scientific and Industrial Research Organisation (CSIRO) has performed population modelling which suggests that repressible-lethal systems may be effective against Northern Pacific Sea Stars (Bax et al. 2006). Oxitec has developed commercially approved mosquitoes with a repressible lethal system that has been effective in commercial field trials in Florida, India, Panama and Brazil(Carvalho et al., 2015).
Sex-specific repressible lethal systems are variants of repressible lethal systems where only males/females die in the absence of the repressor and the surviving sex passes copies of the gene to subsequent generations to skew sex ratios. Ideally, the engineered organisms carry multiple copies of the construct to mitigate the effects of the gene being diluted out over a few generations. A major advantage of this system over non-sex selective repressible lethality is that larval stages can be released into the wild which eliminates the costs of rearing engineered organisms to maturity.
Female lethal systems have been engineered into several insect species and fish (Fu et al., 2010; Concha et al., 2016; Thresher et al. 2014). However, field trials in mosquitoes have shown that males can also be affected by low levels of gene expression from the female lethal construct so that they are insufficiently competitive with wild males (Facchinelli et al., 2013). It should be possible to address this problem but it will still impose some limit on the number of copies of the genetic circuit males can carry. CSIRO’s modelling has shown that male-lethal repressible systems may outperform other repressible lethal approaches (Bax et al., 2006). Engineering the sex-specificity into these systems can be somewhat challenging and will require knowledge about gene expression differences between sexes.
Pheromone Disruption
Starfish communicate with each other using chemical signals dissolved in seawater. These protein pheromones can communicate the presence of danger or help to co-ordinate spawning aggregations. Recent work by Bernard Degnan’s group (Hall et al. 2017) identified genes encoding some of these pheromones. It might be possible to engineer starfish to misexpress these pheromones and induce mal-adaptive behavior in others. For example, starfish constitutively producing alarm pheromones may disperse mating aggregations or be used as repellants to protect sensitive areas. Misexpression of aggregation signals may be exploited to draw starfish away from sensitives areas and into traps. This remains an entirely theoretical approach for starfish at this time, with no empirical data. However, pheromone disruption is an active control method for some insect pests, used to either trap males or to flood the system so that becomes impossible for males to find females.
Synthetic Species
Synthetic Genetic Incompatibility (SGI) is a method to engineer species-like barriers to sexual reproduction where the SGI ‘synthetic species’ population cannot form viable/fertile offspring with wild type organisms (Maselko et al., 2017). Combining SGI with a sex-specific lethal system (SSIMS) would result in a limited in situ amplification of the engineered population in the wild. When an SSIMS organism mates with wild organisms, no offspring survive. However, when SSIMS mate with other SSIMS, only SSIMS males are generated. Modeling data presented at conferences has shown that SSIMS has the potential to be more effective than repressible-lethal alone for invasive carp (Siba Das, Personal Communication), however, it is not known if this would extend to starfish.
Gene Drives
Threshold independent gene drives can be used to spread genes through a population in a way that alters the standard model of inheritance (Burt 2003). Normally, releasing an organism with a single copy of a recessive-lethal gene (lethal when two copies present) will result in the gene’s dilution as it only gets passed on to half of the offspring per reproductive cycle. If two carriers mate, 25 percent of them will be non-viable homozygotes, 25 percent will not carry the gene and 50 percent will be heterozygotes. If the recessive lethal gene is part of a gene-drive system, all offspring receive a copy of the recessive gene and it can spread through all populations connected by gene-flow. Any mating between carriers results in 100 percent non-viable offspring. Gene drive methods are in advanced development in mosquitos as a mechanism to combat malaria (Hammond et al. 2016) and testing is ongoing in rodents (Leitschuh et al. 2018). However, the technique is controversial due to the concerns over uncontrollable negative effects on ecosystems or from a release of gene drive to unintended areas. Other research outlines large technical hurdles facing gene drives, such as inbreeding, poor homing, and development of resistance mutations (Unckless et al. 2018). Overcoming these challenges will need to be coupled with development of gene-drive systems with population percentage thresholds and/or mechanisms for spatial/temporal control.
RISKS & CHALLENGES
Improvement of detection and monitoring of invasives is an urgent priority. Innovation and further development of eDNA techniques could transform current detection efforts. However, while eDNA offers considerable promise for increasing the timeliness and ease in detecting alien species, its application to support quarantine or large-scale invasive species management requires significant development and standardization. Current eDNA methods can suffer from uncertainties in species identification (especially in marine environments), presents a risk of false positives, and could have weak statistical power (leading to overconfidence when no detections are recorded). Nonetheless, the power of eDNA technologies and their adoption at scale will likely become a major focus of invasion science (Riccardi et al. 2017).
The technical challenges to develop genomic biocontrol of marine invasive and irruptive species are significant but not impossible. A significant amount of foundational research and proof of concept will be required prior to field implementation. Specifically, ecological and population genetic research on the target species is essential to determine the feasibility and effectiveness of the various biocontrol approaches. The genetic approaches for eradicating or reducing the impact of invasive rodents are still in their infancy, and biocontrol methods are even less advanced for ocean species; the timeline to develop a comprehensive field trial proposal is estimated to be a at least five years. To accelerate this timeline, proof-of-concept marine biocontrol projects should target systems in which foundational ecological and genomic research has already been completed. For example, COTS biologists are able to predict with a high level of certainty where and when irruptions are likely to begin, making this invasive species a strong candidate for genomic biocontrol (Russ Babcock, personal comment).
Intervening in nature using genetic biocontrol methods is controversial largely due to the nascent state of the field. Because these are still unproven technologies, it is essential that the field moves forward cautiously and obtains social license from the public and policy makers. Any unintended harm that may result from premature implementation of these technologies would result in a more restrictive regulatory and funding environment and further delay the potential benefits of applying synthetic biology to environmental challenges. In Australia several biocontrol agents have been delayed through the approval process, possibly because of high risk-aversion and limited expertise within the regulatory agencies. Marine applications pose an even higher burden of proof due to the shared nature of marine ecosystems.
Many countries require risk assessments to be carried out before research and pilots with biocontrol agents as a means to predict environmental risk. Post-release studies to validate decisions are rarely required by regulators, but where these studies are conducted, provide valuable information for future decision support.
For some species like the Purple Sea Urchin with a long lifespan, it could take a long time for genetic methods to suppress populations. However, combining genetic controls with manual removal may still help to dampen population explosions in the near term. Species such as sea stars, and many other marine species, represent a hypothetically risky option for implementation of genetic engineering, since their young are planktonic and spread worldwide through cargo ship ballast. This uncontrolled movement may have legal ramifications that must be examined, however, the approaches discussed here would have a limited ability to persist for multiple generations (with the exception of gene-drives) and are not likely to impact distant populations. Examining the legal framework regarding the inadvertent movement of transgenic plant pollen and seeds may be illuminating. Ultimately, it is still unclear what impacts genetically modified ocean species will have if released into the wild, accordingly ecological studies and careful testing will be required prior to implementation in natural systems.
The timelines and financial investment required to get from our current level of knowledge to implementing genetic control methods in the wild could be 5-10 years. However, this risk is mitigated by the knowledge that successful implementation of genetic biocontrol programs will provide a forerunner for other efforts with other species, and that foundational research will provide potential benefits for non-genetic biocontrol efforts. Such research will also provide great benefits to other fields such as marine biology, conservation science, and perhaps even human health.
LEADERS
As relatively isolated islands, where invasive species have an oversized impact, the governments of Australia and New Zealand have significant funding programs for national and international research of invasive and irruptive species.
These efforts include developing innovative technologies and tactics for control and eradication of invasive species (NZ Ministry of Business, Innovation and Employment: Catalyst Fund investing in non-transgenic technology) and continuous marine pest biosecurity research and development and the development and validation of assays for detecting marine pests (Australian Marine Pest Sectoral Committee).
The Bill and Melinda Gates and Tata foundations have invested in genetic biocontrol technology for disease vectors. The US Defense Advanced Research Projects Agency (DARPA) has invested in numerous genetic control projects including providing early funding to the Genetic Biocontrol of Rodents (GBIRd) Program, which is investigating the feasibility of, and assessing the social, ethical, and biological risks of, gene-drive modified organisms for eradication of island invasive species. This is a collaborative effort between governments, NGOs, and research universities including: CSIRO, Island Conservation, Landcare Research, North Carolina State University, Texas A&M University, University of Adelaide, and the United States Department of Agriculture (USDA).
REFERENCES
Ballew, Nicholas G., et al. “Invasive lionfish reduce native fish abundance on a regional scale.” Scientific reports 6 (2016): 32169.
Bax, Nicholas, et al. “Marine invasive alien species: a threat to global biodiversity.” Marine policy 27.4 (2003): 313-323.
Burt, Austin. “Site-specific selfish genes as tools for the control and genetic engineering of natural populations.” Proceedings of the Royal Society of London B: Biological Sciences 270.1518 (2003): 921-928.
Byrnes, Jarrett E., Pamela L. Reynolds, and John J. Stachowicz. “Invasions and extinctions reshape coastal marine food webs.” PloS one 2.3 (2007): e295.
Campbell, Karl J., et al. “The next generation of rodent eradications: innovative technologies and tools to improve species specificity and increase their feasibility on islands.” Biological Conservation 185 (2015): 47-58.
Carlton, JAMES T. “13 The scale and ecological consequences of biological invasions in the World’s oceans.” Invasive species and biodiversity management 24 (2001): 195.
Carvalho, Danilo O., et al. “Suppression of a field population of Aedes aegypti in Brazil by sustained release of transgenic male mosquitoes.” PLoS neglected tropical diseases 9.7 (2015): e0003864.
Colnar, Audrey M., and Wayne G. Landis. “Conceptual model development for invasive species and a regional risk assessment case study: The European green crab, Carcinus maenas, at Cherry Point, Washington, USA.” Human and Ecological Risk Assessment 13.1 (2007): 120-155.
De’ath, Glenn, et al. “The 27–year decline of coral cover on the Great Barrier Reef and its causes.” Proceedings of the National Academy of Sciences (2012): 201208909.
Duncombe, Lynda G., and Thomas W. Therriault. “Evaluating trapping as a method to control the European green crab, Carcinus maenas, population at Pipestem Inlet, British Columbia.” Management of Biological Invasions 8.2 (2017): 235-246.
Ficetola, Gentile Francesco, et al. “Species detection using environmental DNA from water samples.” Biology letters 4.4 (2008): 423-425.
Gurevitch, Jessica, and Dianna K. Padilla. “Are invasive species a major cause of extinctions?.“ Trends in ecology & evolution 19.9 (2004): 470-474.
Hall, Michael R., et al. “The crown-of-thorns starfish genome as a guide for biocontrol of this coral reef pest.” Nature544.7649 (2017): 231.
Hammond, Andrew, et al. “A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae.” Nature biotechnology 34.1 (2016): 78.
Huxel, Gary R. “Rapid displacement of native species by invasive species: effects of hybridization.” Biological conservation 89.2 (1999): 143-152.
Jerde CL, Mahon AR, Chadderton WL, Lodge DM (2011) “Sight-Unseen” detection of rare aquatic species using environmental DNA. Conservation Letters 4(2): 150–57. https://doi.org/10.1111/j.1755- 263X.2010.00158.x
Katsanevakis, Stelios, et al. “Impacts of invasive alien marine species on ecosystem services and biodiversity: a pan-European review.” Aquatic Invasions 9.4 (2014): 391-423.
Keesing, John K., et al. “Large-scale laboratory culture of the crown-of-thorns starfish Acanthaster planci (L.)(Echinodermata: Asteroidea).” Aquaculture 157.3-4 (1997): 215-226.
Leitschuh, Caroline M., et al. “Developing gene drive technologies to eradicate invasive rodents from islands.” Journal of Responsible Innovation 5.sup1 (2018): S121-S138.
Lin, Che-Yi, and Yi-Hsien Su. “Genome editing in sea urchin embryos by using a CRISPR/Cas9 system.” Developmental biology 409.2 (2016): 420-428.
Molnar, Jennifer L., et al. “Assessing the global threat of invasive species to marine biodiversity.” Frontiers in Ecology and the Environment 6.9 (2008): 485-492.
Morris, James A. “Invasive lionfish: a guide to control and management.” (2012).
Pearse, John S. “Ecological role of purple sea urchins.” Science 314.5801 (2006): 940-941.
Pimentel, David, Rodolfo Zuniga, and Doug Morrison. “Update on the environmental and economic costs associated with alien-invasive species in the United States.” Ecological economics 52.3 (2005): 273-288.
Pyšek, Petr, and David M. Richardson. “Invasive species, environmental change and management, and health.” Annual review of environment and resources 35 (2010): 25-55.
Rahel, Frank J., and Julian D. Olden. “Assessing the effects of climate change on aquatic invasive species.” Conservation biology 22.3 (2008): 521-533.
Rast, Jonathan P. “Transgenic manipulation of the sea urchin embryo.” Developmental Biology Protocols: Volume II. Humana Press, 2000. 365-373.
Ricciardi, Anthony, et al. “Invasion science: a horizon scan of emerging challenges and opportunities.” Trends in Ecology & Evolution 32.6 (2017): 464-474.
Richardson, David M., et al. “Invasive alien species and global change: a South African perspective.” Invasive species in a changing world (2000): 303-349.
Robson HL, Noble TH, Saunders RJ, Robson SK, Burrows DW, Jerry DR (2016) Fine tuning for the tropics: application of eDNA technology for invasive fish detection in tropical freshwater ecosystems. Molecular Ecology Resources 16: 922–932, https://doi.org/10.1111/1755-0998.12505
Ross, D. J., et al. “Interaction and impacts of two introduced species on a soft-sediment marine assemblage in SE Tasmania.” Marine Biology 144.4 (2004): 747-756.
Roughgarden, Jonathan, and Fraser Smith. “Why fisheries collapse and what to do about it.” Proceedings of the National Academy of Sciences 93.10 (1996): 5078-5083.
Sodergren, Erica, et al. “The genome of the sea urchin Strongylocentrotus purpuratus.” Science 314.5801 (2006): 941-952.
Stepicheva, Nadezda A., and Jia L. Song. “High throughput microinjections of sea urchin zygotes.” Journal of visualized experiments: JoVE 83 (2014).
Sutherland, William J., et al. “A 2017 horizon scan of emerging issues for global conservation and biological diversity.” Trends in Ecology & Evolution 32.1 (2017): 31-40.
Thresher, Ronald E., et al. “Genetic control of invasive fish: technological options and its role in integrated pest management.” Biological invasions 16.6 (2014): 1201-1216.
Unckless, Robert L., Andrew G. Clark, and Philipp W. Messer. “Evolution of resistance against CRISPR/Cas9 gene drive.” Genetics 205.2 (2017): 827-841.
Uthicke, S., et al. “Outbreak of coral-eating Crown-of-Thorns creates continuous cloud of larvae over 320 km of the Great Barrier Reef.” Scientific reports 5 (2015): 16885.
Zaiko, Anastasija, et al. “Metabarcoding approach for the ballast water surveillance–an advantageous solution or an awkward challenge?.“ Marine Pollution Bulletin 92.1-2 (2015): 25-34.





Back to Top