Big Ideas
CONTEXT
Coastal fishing nations stand at the frontline in the battle against illegal fishing (Agnew et al. 2009). Recent multilateral agreements concerning illegal, unreported, and unregulated fishing (UN Agreement on Port State Measures 2018) highlight the need to develop tools for accurate and cost-effective monitoring, control, and surveillance of fisheries. Conventional tools are limited because they are time-consuming and require expertise in fish identification (Mora et al. 2009). Many developing countries report monitoring capacity to be one of their major hurdles in improving fisheries management (Morgan et al. 2007), including a deficiency in hands-on training and local technical capacity. Current methods are ill-equipped to accurately and efficiently distinguish among the tremendous biodiversity within fish landings. Immediate innovations to improve seafood traceability and long-term sustainability are critically needed.
Thailand is one country with a particularly acute need for innovation in the fisheries management sector. Tens of thousands of Thai people depend on the seas for their livelihood yet, Thai fishery stocks are over-exploited. The over-capacity of the fishing industry in the Gulf of Thailand ties coastal populations to harvesting needs, including through the non-selective practice of capturing ‘trash fish,’ fish generally not considered suitable for human consumption but used for fish meal, animal feed, or fertilizer. Included in these landings are juveniles of valued fishes (some estimates 30% of catch), yet accurate identification of which fish are over-exploited is limited.
Thai fisheries regulation, such as the Royal Ordinance on Fisheries B.E. 2558 (2015), provides both the framework and resources for improving coastal fisheries, but reaching such goals requires building local capacity in effective, rapid fish identification technology that will help guide management decision on gear-use, fishing locations, and biodiversity and biological insight into Thai fish populations. This project directly targets the needed modernization of fisheries monitoring and can also facilitate Thai seafood sector’s compliance with stringent, recently implemented import regulations in the U.S. and E.U. (Willette and Cheng 2018).
Emerging genetic and satellite-based technologies provide a new opportunity to solve this long-standing monitoring challenge as well as creating new opportunities for accurate fish identification (Willette et al. 2014). For example, DNA barcoding is increasingly used to detect seafood mislabeling in sushi restaurants and grocery stores (Willette et al. 2017, Willette et al. 2018), and now to identify marine fish biodiversity from environmental DNA (eDNA) in seawater (Kelly et al. 2014). While additional indexing and barcoding are needed, we can now leverage existing and free databases of genetic information (e.g. Barcode of Life project, Janzen et al. 2005) to enable the rapid identification of commodity fishes versus rare or threatened fish species.
This project develops and implements a non-invasive genetics technology to determine the composition of targeted catch and by-catch from fishing vessel wastewater. Fish readily shed DNA into the environment, creating eDNA that can be sampled and identified to species level through genetic barcoding. Importantly, shed DNA begins to degrade after 72 hours, thus this targeting the wastewater from fishing vessels at port will provide a profile of the fish contained in the present landing, not past catches or from the adjacent waters.
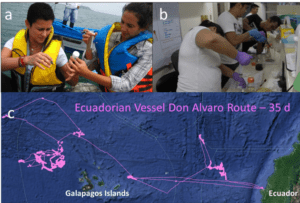
Figure 1 – Preliminary field, lab, and vessel tracking: a) Field sampling from artisanal vessel in Ecuador; b) eDNA library preparation by researchers in Philippines; and c) Vessel-tracking by GFW (pink line) of Ecuadorian ship Don Alvaro prior to July 2017 sampling in Manta, Ecuador.
Furthermore, this project will utilize free and public data from satellite-based automatic ship identification systems (AIS) to track fishing vessel movement worldwide. AIS is a type of on-board transponder that publicly broadcast’s a vessel’s identity and position and can be paired with satellite technology that allows mass detection of AIS dispatches (McCauley et al. 2016). Organizations such as Global Fishing Watch (globalfishingwatch.org) have developed algorithms using the broadcasted AIS data to scrutinize the transit patterns of fishing vessels and generate global maps of where and when fishing vessels fish (Jablonicky et al. 2016, Kroodsma et al. 2017). Vessel data is reported in near-real time, is publicly and freely available, and can provide a port-to-port transit map of monitored fishing vessels (see Figure 1c for example).
Screening the composition of every landing from every vessel is impractical and cost-prohibitive. This project aims to narrow the monitoring focus on vessels with suspicious fishing behavior. Fishing activity is distinguishable in ‘zig-zag’ routes and slowed vessel speeds can be identified via satellite-based tracking. Fishing activity in no-fish areas, for example, can be observed and inform regulators which vessels may warrant screening, even prior to the vessel’s arrival to port. By developing an understanding of what fish and bycatch are caught on a particular vessel’s route, fisheries managers can significantly improve spatial and temporal regulations.
Preliminary field and laboratory eDNA method testing and use of AIS vessel data were completed in summer 2017 in collaboration with academic and regulatory partners in Ecuador and the Philippines (Figure 1). This development phase showed great promise for an expanded application.
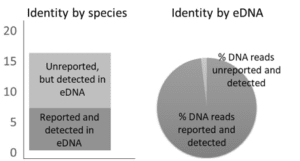
Figure 2
Results from Preliminary Field and Lab Testing
Successful field sampling, lab work, and bioinformatics have been completed as designed, producing a profile from fish landings. For example, nearly 200,000 high-quality DNA reads were obtained from triplicate wastewater sampling of the Ecuadorian vessel Don Alvaro in July 2017. This vessel’s fishing route was tracked during its 35-day voyage prior to arrival in Manta, Ecuador (Figure 1). Results show that only 7 of the 16 species identified by our method were reported by the vessel (Figure 2), those species not reported made up only a small fraction (<2%) of sequenced reads and were identified as typical by-catch species. Figure 2 compares the reported and DNA-identified fish species by Ecuadorian vessel Don Alvaro from wastewater samples.
BIG IDEA
Designed in close collaboration with local Thai researchers, this project will deliver powerful and accessible tools and the necessary hands-on training at the user-end to demonstrate the power of coupling eDNA collection and analysis with satellite tracking of fishing vessels in Thailand. These tools will be freely transferable to fisheries stakeholders world-wide, extending the promise of this approach to initiate a dramatic global transformation in fisheries monitoring.
This project leverages hands-on training and collaborative research to deploy and extensively test and validate cutting-edge tools to modernize monitoring of fish landings. This project has the broader targeted benefits to:
- Foster constructive collaboration among fishery stakeholders, students, and scientists in promoting evidence-based decision-making with powerful, accessible tools;
- Increase awareness and build global research capacity in how to engage in and solve current limitations in coastal fisheries monitoring and management;
- Evaluate capture-fishery for the presence and frequency of key ecological species in landings and grow our understanding of their distribution and role in healthy systems; and,
- Develop a deeper understanding of fishing effort with the utilization/exploitation of certain ecologically valuable species.
Experimental Design and Implementation
This field pilot study will use publicly-available AIS data to track and profile the behavior of Thai fishing vessels and use eDNA methods to identify fish landings (including bycatch) to species-level accuracy across time and location. By comparing genetic data to vessel-reported fish manifests, we will create vessel-level and port-level fish identity rates, and demonstrate the feasibility and application of a versatile, powerful, and accurate monitoring method at the beginning of the global seafood supply chain.
Field work will initially occur at Songkhla Port, with additional sites added on in subsequent years of the project. Songkhla Port is the primary fishing port in southern Thailand along the Gulf of Thailand, and whose proximity (~30km) to project collaborators at Prince of Songkhla University (PSU) make it the ideal field site for monitoring. In close collaboration with our Thai partners, triplicate landing wastewater samples will be taken from 10 representative fishing vessels in each of 6-month field seasons for a total of 12 months (n = 360) and screened for fish composition. Selection of vessels will be made with local partner considerations who have already established relationships with regional fishers and by using the public database maintained by Global Fishing Watch (www.globalfishingwatch.org).
Satellite-based tracking data will be used to re-construct the fishing route of the 10 vessels just prior to sampling. Briefly, the identity and maritime number of each sampled fishing vessel at port will be recorded for subsequent route reconstruction using the Global Fishing Watch database. All vessels’ routes will be mapped and location of where fishing activity took place will be identified by securitizing vessel cruising speeds (< 4 knots suggests fishing) and occurrence of ‘zig-zag’ routes. Fishing hotspots (areas of high fishing effort) will be identified from aggregating routes from the 10 vessels over 12 months, as will instances of fishing activity in violation of national and international law. Tentatively, it may be possible to infer geo-location of key marine species by overlaying fishing vessel maps with eDNA species identity data.
Wet lab processing and preparation of eDNA libraries will be conducted as described in methods developed in Ecuador in 2017 and freely available by request at willettelab.com. Specifically, individual wastewater samples will be filtered to separate tissue cells, extracted for genomic data, and sequenced using universal primers (targeting the 12s gene region) designed for 880 fish species (Miya et al. 2015) to target a broad range of fish and elasmobranch DNA. To strengthen confidence in profiling species biodiversity, a second gene region (Cytochrome oxidase I – COI gene) will be targeted from all eDNA samples to cross-validate 12S results. Massive parallel sequencing of samples on the Illumina platform will produce millions of raw genetic reads per sample, with bioinformatics on samples using the ranacapa pipeline, a recently developed R-based package for analyzing and visualizing eDNA sequencing results (Kandliker et al. 2018). High quality reads will be compared to the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) to identify harvested species. Summary statistics will be calculated and used to estimate vessel-level and an aggregated port-level fish identity rates in comparison to landing manifests, resulting in an unprecedented report of fish landing biodiversity unique to each sample fishing vessel.
PARTNERS
Project Management – Demian Willette (Loyola Marymount University – LMU)
eDNA Library Preparations – Demian Willette (LMU, lead), U.S. & Thai Project Participants
Training – Demian Willette (LMU), Anchana Prachep (Prince of Songkhla University – PSU)
Project Location – Thailand – Port of Songkhla and Prince of Songkhla University Hat Yai
Implementation – Demian Willette (LMU), Anchana Prachep (PSU), James True (PSU)
BUDGET
| ITEM | USD | DETAILS |
|---|---|---|
| Training Workshops | $40,000 | 2-week course and hands-on intensive training in eDNA methodology & bioinformatics, managing satellite vessel data, and in-field sampling training at Songkhla Port. Includes flight and per diem for D. Willette, post doc, A. Prachep, and J. True. $20,000 per workshop for two years. |
| Thai Participation Support | $18,000 | $60 x 360 eDNA library preparations; lab plastic/reagent consumables; 1 thermal cycler; 1 centrifuge; 2 pipette sets. |
| Reagents & Supplies | $35,000 | $3,500/lane x 6 lanes (60 preps per lane). |
| eDNA Sequencing | $21,000 | Collection and sampling kits for obtaining voucher/tissue specimens (up to 100 specimens) for improving 12S/COI genetic database, round-trip shipment costs, fees for export/import of tissues. |
| Tissue Collection | $5,000 | Collection and sampling kits for obtaining voucher/tissue specimens (up to 100 specimens) for improving 12S/COI genetic database, round-trip shipment costs, fees for export/import of tissues. |
| Synthesis Meeting | $20,000 | 2-day meeting in Hat Yai, Thailand. Includes venue, flights, accommodations, and meals for project collaborators, project participants, and invited stakeholders from Thai and regional governments, NGOs, and academia. (Travel/sustenance averages $300 for 15 loca participants and $1000 for 8 international participants). |
| Post Doc Researcher | $140,000 | 2-year position: Salary + fringe benefits; hired through competitive application; responsible for supporting workshops and 6-month extended support of Thai project participants at PSU |
| PI Summer Salary | $67,500 | Calculated on AY base salary + fringe benefits for 3 months per year, for 2 years. |
| Indirect Costs | $34,650 | 10% research overhead. |
| Total: | $381,150 |
RISKS & CHALLENGES
- Species sourcing and existing genetic database: This experimental design addresses the anticipated real-world obstacle of working with fish landed from multiple locations in a dynamic fish port in two ways: a) detailed record-taking of fish and fishing location data from the source vessels in the local language in collaboration with our PSU partners; and b) building database of local and regional fish tissue samples and genetic reference data. Notably, the existing publicly available COI database has gaps in species for Southeast Asia. Leveraging past collaborations from around Southeast Asia, we will obtain both voucher museum specimens and fresh-caught specimens from across the Indo-Pacific region. The resulting genetic database will be a valuable, openly shared resource for researchers that will last long beyond the duration of this project.
- Avoiding unintended economic hardship: Ensuring sustainable livelihoods of fishing communities and the broader seafood sector requires careful consideration of unique governance and socio-economic context in each fishery and nation (Challender et al. 2015). I have worked with fisheries stakeholders across nine Indo-Pacific nations, and this understanding of local to global fisheries challenges is inherent in this proposal’s partnerships and co-development.
- Technology transfer: To support meaningful and sustained implementation of these technologies, this project will to produce a tool and resulting data that is transferable, easy-to-use, and highly accessible to fisheries scientists and regulatory agencies around the world. Importantly, through my diverse network of collaborators and availability of these methodologies on my website, these resources will be publicly available for anyone to utilize and build upon in our shared commitment to fisheries sustainability.
- Building research capacity: Currently, many Pacific developing countries report monitoring capacity to be one of their major hurdles in improving fisheries management (Morgan et al. 2007). Thus, for initiatives combating illegal and unregulated fishing to succeed, fisheries monitoring data must be made more efficient, including by optimizing methods like this eDNA tool. Hands-on experience is the most effective and often most efficient way to gain knowledge and understanding of new topics and methods. To this end, I will lead a two-week mini-course and training in each year, training 4-6 Thai participants (graduate students & resource managers recruited with Dr. Anchana Prachep – PSU), who will received sustained training and support for an additional 6 months by the visiting project post-doc. Importantly, travel and a modest stipend will be provided to Thai participants to reduce barriers to participation.
TIMELINE
The proposed timeline of project activities occurs on an annual cycle in consideration with PI Willette’s full time assistant professor responsibilities and duties during the academic year at Loyola Marymount University, and as planned with international collaborators. Research activities occur over 6 months each year with mini-course and intensive training in May – July, and sustained support and training August – October.
1. Pre-Production (6 Months)
- Draft and sign MOU between LMU and PSU
- Obtain necessary permits and documents for research in Thailand
- Purchase research supplies
- Detailed planning of field season with Thai collaborators, schedule flights and fieldwork
- Recruitment of Thai participants through a short application process with PSU
MILESTONES:
a) Agree upon and sign MOU (LMU and PSU)
b) Sign and obtain research permits
c) Purchase and obtain research supplies
d) Recruit and plan transportation of 4-6 Thai project participants
e) Schedule, book, and finalize travel, accommodations, and transports for field work
2. Pilot: Workshop & Field Implementation (6 Months)
- Launch project with two-week mini-course and hands-on intensive training at PSU with local collaborators and 4-6 project participants.
- Conduct first round of fishing vessel eDNA sampling & wet lab library preparations
- Send eDNA library preparations for sequencing.
- Training participants in bioinformatics using existing data from Ecuador samples.
- 6-month sustained support of participants by visiting project post-doc at PSU.
- Re-create vessel routes from satellite-based tracking for each of 10 vessels per month.
- Generate a ‘fishing hotspot’ map from aggregate data of 10 fishing vessels over 12 months.
MILESTONES:
a) Complete two-week mini-course and hands-on intensive training in the molecular lab space at PSU with participants, inclusive of field experiences and bioinformatics training.
b) Complete 120 sampling events (triplicates from each vessel, n = 360).
c) Complete wet lab library preparations on 360 samples, send for sequencing.
d) Implement bioinformatics pipeline on 360 samples, produce species identity based on 12S and COI genetic identifications.
e) Complete individual and aggregate fishing vessel maps and identify fishing hotspots.
3. Evaluation & Reporting (Ongoing)
- Pilot success will be assessed per metrics stated above.
- Annual reports will be drafted in collaboration among U.S. and Thai partners and participants and released in accordance with agreed upon terms in MOUs.
MILESTONES:
a)
4. Synthesis Meeting (2 Weeks)
- A cumulating analysis of the project’s results and findings will occur in late 2021 after the second field implementation season, with invitations extended to Thai and regional fisheries stakeholders including governments, NGOs, and academics.
MILESTONES:
a) Successfully plan and conduct synthesis meeting with stakeholders at PSU in late 2021.
b) Assess project participants’ comfort and confidence in implementing eDNA and satellite vessel tracking approach through presentations during synthesis meeting, and tentatively peer reviewed scientific publications.
c) Dissemination of the ground-truth methodology developed in this project to stakeholders freely via willettelab.com website and through regional channels with stakeholders.




